Artificial Intelligence and deep learning models for biochemical applications
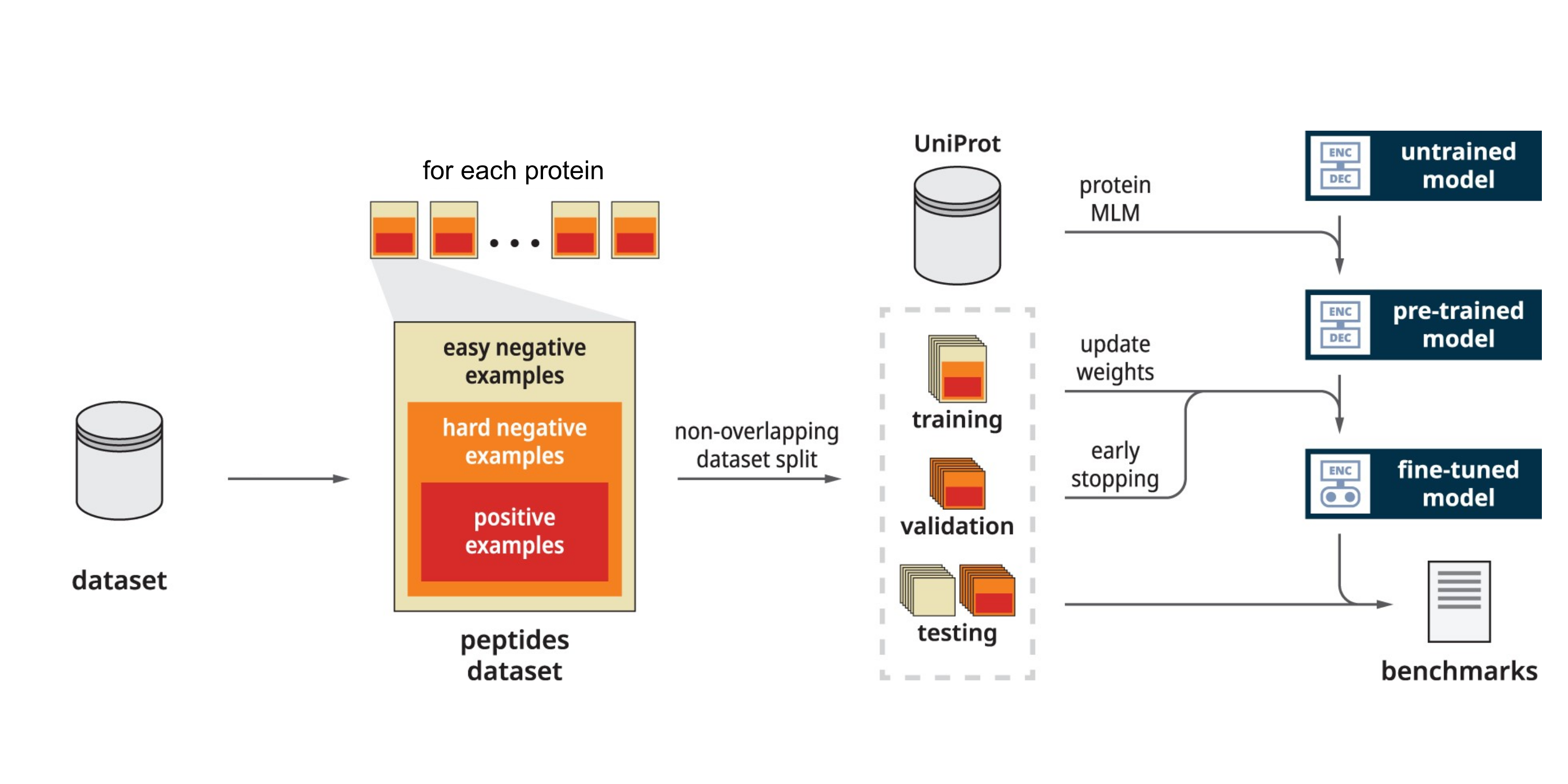
The Kannan lab is exploring cutting-edge AI research in the field of protein function prediction. One area is understanding and predicting post-translational modifications, which are critical to regulating cellular processes. We developed a unified framework (Phosphormer) [1] using protein language models to generate context-aware features from primary protein sequence data to predict kinase-specific phosphosites. We are working on enabling this model to make accurate phosphosite predictions across the entire kinome and expand to other post-translational modifications. We have further developed workflows and visualization methods to classify protein families using sequence embeddings or feature-rich numerical representations of protein sequences. These embeddings infer proteins' structural and functional properties based solely on sequence information. We successfully applied this in the classification of phosphatases, protein kinases, and the S-Adenosyl-L-Methionine (SAM) enzyme superfamily, generating Neighbor-Joining (NJ) embedding trees as an orthogonal method to phylogeny for visualization of evolutionary information [2, 3, 4].